

| Home | Search | Browse | Statistics | User guide | FAQs | Links | Questions | Contribute | Download |
Frequently Asked Questions (FAQs)Uterus is a major female reproductive tissue. The available gene expression databases provide very less or no information for the normal and abnormal uterine conditions. There are few databases which are either specific to a sub-tissue of uterus or condition. Hence, MGEx-Udb was developed by using a novel meta-analysis approach (BMC Genomics. 2010 Aug 11;11:467) that involves manual curation of maximum data, derived from large-scale gene expression studies, collected from major repositories and publications. It covers a wide variety of physiological, pathological and experimental conditions, pertaining to mammalian uterus and thus serves as a comprehensive resource for the scientific community. A 'reliability-score' for each gene along with the expression status is provided, which indicates the level of agreement across studies. A 'reliability-score' for each gene, under a specific condition(s) and/or location(s) is derived by the extent of agreement or contradiction for each gene's expression, across comparable gene lists (degree of consistency). Hence, the higher the reliability scores, the higher the association with the chosen condition. MGEx-Udb primarily provides a list of genes transcribed or dormant in several normal/pathological physiology and other experimental conditions pertaining to mammalian uterus, including the derived reliability score. In addition, MGEx-Udb provides basic gene, transcript, promoter, protein details, Gene Ontology terms, protein-protein interactions and relevant PubMed articles for the genes. Expression status based on EST (from UniGene) and curated next generation sequencing data is also provided for each gene. Refer user guide for more information on the results and output features. We recommend the use of 'Mozilla Firefox 3.0 or IE 8.0' for our database. Please enable javascript in your browser. The database works reasonably well with other browsers too. However, a few specific features may not be functioning optimally. MGEx-Udb covers huge amount of gene expression data, mostly large-scale, from available publications and various gene expression repositories such as GEO, ArrayExpress, and caARRAY. Majority of the data comes from microarray studies, and remaining from proteomic and small-scale mRNA studies. RNA Sequencing (NGS) data is collected from published literature to provide an indicative expression status of the genes. Whereas, EST based expression information is taken from UniGene. Yes. If you are interested in downloading the complete data, please send a request to kshitish@ibab.ac.in. Yes, there is an option provided in the result page, to download the lists in a text format. Refer user guide for more information on the results and output features. Yes, for each gene-set (transcribed/dormant list of genes) in the results, you are also provided with a list of references from where the data is curated. Yes. The contribute section of the database allows the user to upload the data, after completing a simple registration form. Currently, the database covers human, mouse, rat, cow and pig. We intend to update the database with data for other mammalian species as well. May be normal is assigned to those gene-lists where, the tissue/cell-line/cells considered for profiling is histologically normal, but has undergone some control treatments or is taken from an individual who has other disorder(s) or undergone a surgery. In other words, these conditions are not considered as completely normal. May be normal is divided in to various types such as vehicle treated, sham surgery, dietary alterations, and patient, depending on the reason for which they were considered as may be normal. For example, "May be normal - patient" for cervix tissue, corresponds to a condition where, histologically normal cervix tissue of an individual who has other disorder (e.g., endometriosis) is considered for profiling. Control is assigned to those gene-lists where, the tissue/cell-line/cells considered for profiling is from any physiological/experimental condition (except normal), that is control treated. Sub-types under control correspond to the type of control treatment. For example, "Disease - control" for uterus tissue, corresponds to a condition where, a diseased uterus tissue has undergone a control treatment (e.g., vehicle treatment, control vector treatment, etc.). Specific gene study is assigned to those gene-lists where, the tissue/cell-line/cells were used to study gene effects either by transfection, mutation or knockout. This condition is assigned to those gene-lists where, the experiments involved in-vitro culturing of either primary cells or isolated cells. There are few conditions (mostly experimental), which has lesser studies compared to the major conditions such as normal, disease, hormone treatment. Such conditions are assigned to "others" category, which is further classified based on the actual condition (e.g., gestation, growth factor treatment, post-parturition). This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, has genetic aberrations such as deletions/insertions. It also refers to the zygosity of a species. This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, is cultured with a medium enriched or depleted with specific components (e.g., human trophoblasts). This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, is known to be cultured in a standard/alternative growth medium (e.g., growth factor deprivation). This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, is known to be resistant to a specific drug (e.g., cisplatin). This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, is known to be either grown or cultured in a standard/specific environmental condition (e.g., hypoxia). This condition is assigned to those gene-lists where, the endometrium/endometrial-cells used for profiling, is taken after transferring or implanting an embryo. This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, is known to have a specific (e.g., positive/negative) response to therapy (radiation/surgery). This condition is assigned to those gene-lists where, the tissue/cell-line/cells used for profiling, taken from a subject known to have a specific habit (smoking/non-smoking). This condition is assigned to those gene-lists where, the cell-line/cells used for profiling, is treated with lipopolysaccharide (LPS). This condition is assigned to those gene-lists where, an abnormal tissue/cells (disease/treated) used for profiling, is taken from a subject suffering with another disorder. This condition is assigned to those gene-lists where, tissue/cells used for profiling, is known to have undergone any surgical treatment earlier. A controlled vocabulary was set for the various normal-physiological and pathological conditions (including experimental conditions) to maintain the uniformity while collecting and uploading the data. The same hierarchy was provided to query those data-sets (currently enabled up to 4 levels). An example hierarchy of a condition and its sub-conditions is shown in the figure given below. The figure highlights hierarchy for "stage IIA non-keratinizing squamous cell cervical carcinoma". 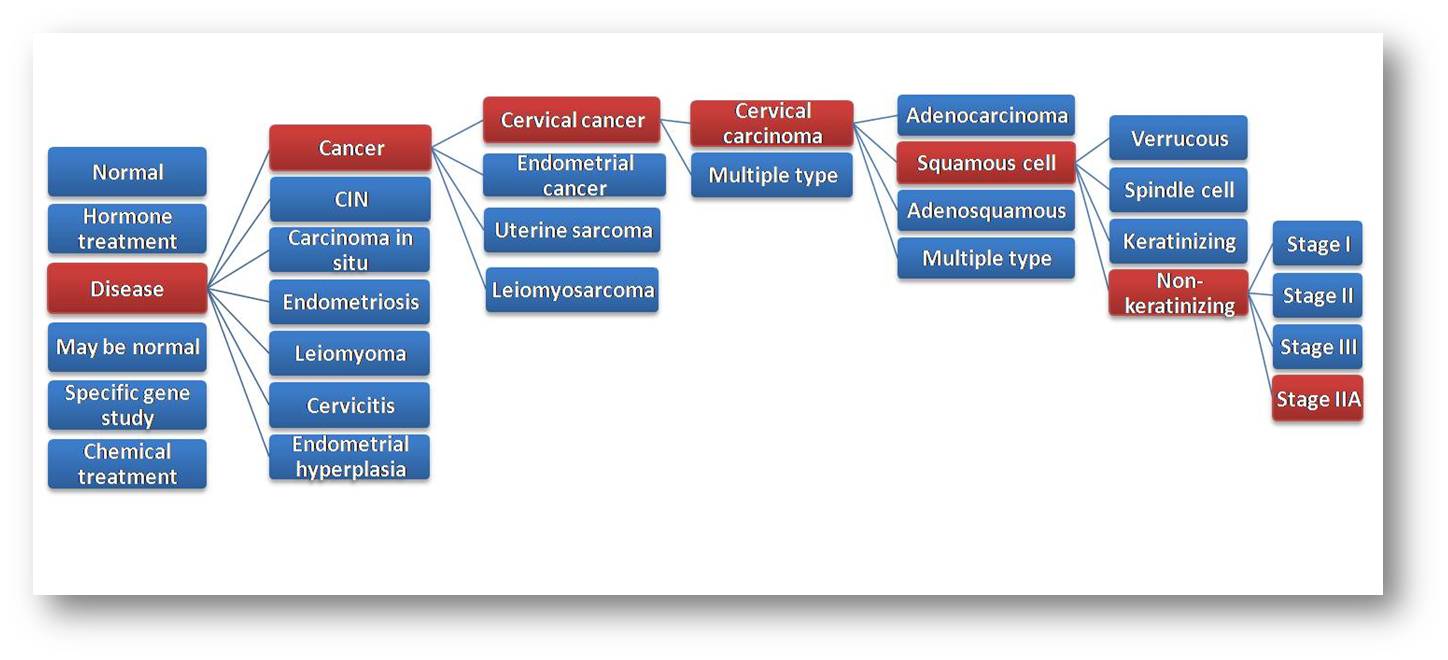
A search strategy was carefully designed to collect relevant articles reported in the literature (detailed procedure can be found at http://dx.doi.org/10.1038/npre.2011.2101.3). Briefly, this involved identifying combinations of query-terms/phrases (see below for more detailed information on query terms or phrases used) for each search tool, obtaining the citations using multiple tools and then compiling the hits into a union list using the Citation-Compiler tool(http://www.shodhaka.com/compiler). More details..
"Data source" provides the references (studies) from where the gene lists are collected and used for derivation of consensus expression pattern of genes for the queried condition. Each genes' expression pattern and score may not be derived from all of the studies listed under "data source" (since all the studies may not cover same genes). If you use MGEx-Udb, please cite the following article: Akhilesh K. Bajpai, Sravanthi Davuluri, Darshan S. Chandrashekar, Selvarajan Ilakya, Mahalakshmi Dinakaran, Kshitish K. Acharya. (2012) MGEx-Udb: A Mammalian Uterus Database for Expression-Based Cataloguing of Genes across Conditions, Including Endometriosis and Cervical Cancer. PLoS ONE. 7(5): e36776. doi:10.1371/journal.pone.0036776 |